Research
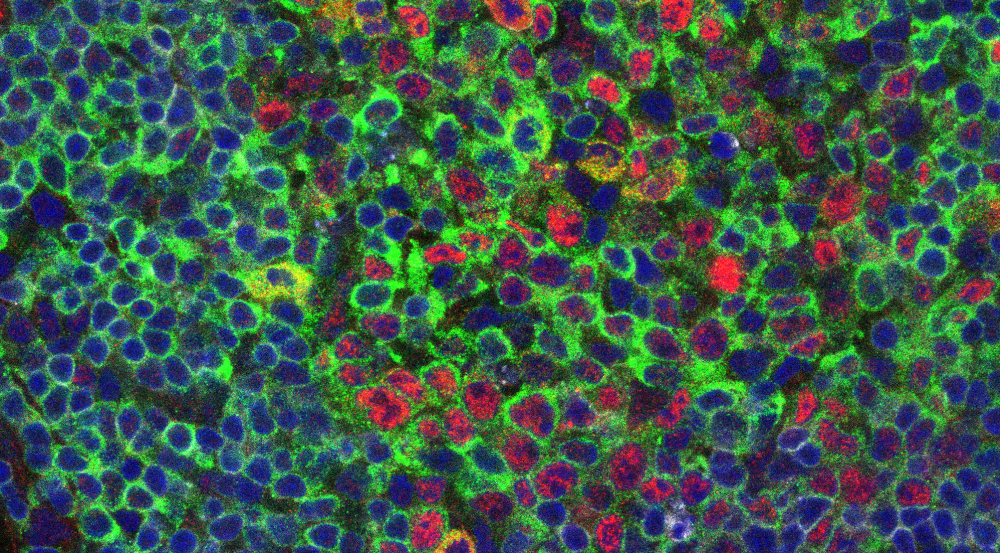
Image: Hyunju Oh-Strauß: Confocal immunofluorescence of c-Rel nuclear presence in Germinal Center B Cells
Overview
Our immune system protects us against invasion by foreign pathogens, such as bacteria, viruses and parasites. However, we pay an evolutionary price for our highly efficient and sophisticated immune defense, namely immunopathology. Immunopathologies include exaggerated responses to harmless substances, also termed allergic responses, misguided responses against our own body, which can lead to autoimmune diseases and the uncontrolled expansion of immune cells, which in turn can cause cancer.
We investigate the molecular and cellular mechanisms underlying such disorders. Immune cells can identify foreign microbial components through a host of cell surface receptors. These receptors then relay signals to the nucleus, where transcription factors activate the expression of genes whose protein products help fight the invaders. Misguidance of such signal transduction events can result in autoimmunity and leukemia or lymphomas, the most prevalent cancers of the immune system.
To study critical mechanisms of immunopathology, we employ
- Genetic loss and gain of function approaches in the mouse
- Cellular and biochemical approaches
Within the immune systems, the main focus is on B and T lymphocytes and mast cells. In these cells, we investigate the regulation of genes and proteins whose exaggerated functions or malfunctions directly contribute to immunpathologies.
In the context of NF-kB signal transduction we currently focus on the ubiquitin editing enzyme A20/TNFAIP3 and the transcription factor c-Rel. Another central topic is the regulation of mRNA stability by the RING finger proteins Roquin1/2. In addition, we are investigating the role of T cell receptor-expression and signaling on immune-modulating regulatory T (TR) cells and lipid-recognizing NK-like T (NKT) cells.
Research Projects
Overview
In this project we investigate the development and functional differentiation of NKT cells. We established a genetic system which allows the generation of a wave of synchronised developing NKT cells in vivo. We employ this system to study how signaling, transcription factor dynamics, anatomical location and cellular contacts influence gene expression and thereby the differentiation of NKT cells into distinct functional subsets, which shape and control T cell responses.
More information: https://www.sfb1054.med.uni-muenchen.de/index.html
Publications related to the project
Dashtsoodol N, Bortoluzzi S, Schmidt-Supprian M. T Cell Receptor Expression Timing and Signal Strength in the Functional Differentiation of Invariant Natural Killer T Cells. Front Immunol. 2019 Apr 26;10:841. doi: 10.3389/fimmu.2019.00841 (link is external).
Drees C, Vahl JC, Bortoluzzi S, Heger KD, Fischer JC, Wunderlich FT, Peschel C, Schmidt-Supprian M. Roquin Paralogs Differentially Regulate Functional NKT Cell Subsets. J Immunol. 2017 Apr 1;198(7):2747-2759. doi: 10.4049/jimmunol.1601732 (link is external).
Vahl JC, Heger K, Knies N, Hein MY, Boon L, Yagita H, Polic B, Schmidt-Supprian M. NKT cell-TCR expression activates conventional T cells in vivo, but is largely dispensable for mature NKT cell biology. PLoS Biol. 2013;11(6):e1001589. doi: 10.1371/journal.pbio.1001589 (link is external)
Haematopoietic stem cells are embedded in the semi-solid tissue of the bone marrow where they give rise to all different types of blood and immune cells in a process called haematopoiesis. This complex cell differentiation process is scrutinized and fostered by a diverse group of bone marrow cells which provides a supportive microenvironment or haematopoietic niche. Stromal cells, for example, are known to present and secrete signaling molecules and stimulating factors like CD40, CXCL12 and IL-7 in order to modulate normal haematopoiesis. Haematological malignancies were previously thought to be driven solely by genetic or epigenetic lesions within the haematopoietic cells. However, based on whole genome sequencing and genome-wide association studies the bone marrow microenvironment has being recognized as having a central role in the pathogenesis and chemoresistance of haematological malignancies. Specific alterations and aberrant signaling networks in the niche can serve as predisposition events that permit mutant haematopoietic cell survival and expansion at early stages of leukemic transformation. Moreover, at later stages during malignancy progression bone marrow niche cells play a central protective role as malignant cells are shielded from chemotherapy leading to ineffective drug treatments and high risk of disease relapse.
Our aim is to elucidate molecular mechanisms underlying the bidirectional communication between haematopoietic niche cells and malignant immune cells in the bone marrow. Through our in vitro and in vivo experiments using proteomics and functional genomics we gain molecular clues and insights into signaling networks pivotal to the cytoprotective microenvironment and therefor to disease manifestation.
Understanding the composition and function of haematopoietic niches during health and disease will help us to identify new potential targets for treatment of haematological malignancies in the future.
More Information: https://web.med.tum.de/en/for2033/homepage/(link is external)
Overview
Despite the recent development of novel therapies lymphomas remain a significant source of human morbidity and mortality, largely due to the clonal evolution of resistant cells. The spectacular success of next generation sequencing tremendously enhanced our knowledge of the identity and prevalence of recurrent, putatively oncogenic alterations and allowed inferences regarding the temporal sequence of mutation acquisition. In contrast, model systems to manipulate, monitor and investigate clonal evolution of lymphomas in vivo are lagging behind. To monitor and investigate the role of clonal competition in indolent malignancies, we propose to address the following specific aims:
Aim 1: To establish and validate genetic tools allowing the introduction and simultaneously the cellular tracking of defined genetic alterations in mouse models for human lymphoma
Aim 2: To assess the expansion, homeostasis and transcriptional/epigenetic changes of follicular lymphoma clones carrying defined genetic alterations.
More information: https://www.sfb1243.biologie.uni-muenchen.de/index.html(link is external)
Overview
Antibodies secreted by B cells of the adaptive immune system establish an essential barrier against bacteria and viruses and their presence is the hallmark of protective vaccinations. B cells are licensed for their tasks during germinal center (GC) reactions and differentiation into antibody-secreting plasma cells. Unfortunately, B cell-derived autoantibodies and proinflammatory cytokines can cause or contribute to autoimmune diseases.
While major transcription factor networks regulating protective (or pathogenic) GCB cell responses have been identified and characterized, little is known about the post-transcriptional regulation by RNA-binding proteins (RBP), whose number rivals that of transcription factors.
We postulate that RBPs exercise critical post-transcriptional control over germinal center B (GCB) and plasmacytic cell physiology and we aim to identify and molecularly characterize these regulatory mechanisms.
To date, we extended and optimized plasmablast generation in the induced germinal center B cell (iGB) culture system, which recapitulates key aspects of plasma cell differentiation in the mouse in vitro. These cells can be efficiently infected with lentiviral and retroviral vectors for genome and gene expression manipulation. We continue to optimize various aspects of the iGB cell system for screens and RBP functional analyses. We built a large repertoire of hematopoietic progenitor cell pools (Hoxb8FL) of different genotypes and are actively optimizing Hoxb8FL differentiation protocols into B lineage cells in vitro and in vivo to the germinal center and plasma cell states. Furthermore, we established various gene editing protocols in these cells and are validating in vivo differentiation potential after each manipulation. We obtained a retroviral libary to globally screen for RBP functions. We established an additional system to expand hematopoietic precursor cells in vitro for adoptive transfer in vivo that does not require expression of exogenous transgenes. We made progress in determining the roles of individual RBPs in terminal B cell differentiation. For validation purposes, we established and characterized a collection of mouse B cell lines resembling mature B cells, B cells bearing features of germinal center B cells and plasma cells. We established genome manipulation protocols in these cell lines and in vivo transfer.
We expect to pinpoint critical roles for RBPs during germinal center reactions and plasmacytic differentiation. Furthermore, we aim to identify RPBs binding to 3prime UTRs of the mRNA of critical mediators of germinal center reactions and plasmacytic differentiation by mass-spectrometry based proteomics. We want to uncover the functional significance of the underlying regulation.
Overview
The transcription factor c-Rel, central to innate and adaptive immune responses, is directly implicated in oncogenesis. However, the mechanisms by which c-Rel alterations drive human hematopoietic and solid cancers remain unresolved. To address this issue, we established mice that allow for cell type-specific enhanced c-Rel expression and visualization. We will employ these and complementary loss of function models as well as advanced proteomic approaches to define tumor cell-intrinsic roles of c-Rel signaling in lymphoma and colorectal cancer, as well as mechanisms and consequences of c-Rel activity within the tumor immune environment.
More information: http://www.sfb1335.med.tum.de/en(link is external)
Publications related to the project
Kober-Hasslacher, M., & Schmidt-Supprian, M. (2019). The Unsolved Puzzle of c-Rel in B Cell Lymphoma. Cancers, 11(7), 941. https://doi.org/10.3390/cancers11070941 (link is external)
Kober-Hasslacher M, Oh-Strauß H, Kumar D, Soberón V, Diehl C, Lech M, Engleitner T, Katab E, Fernandez Saiz V, Piontek G, Li H, Menze B, Ziegenhain C, Enard W, Rad R, Böttcher JP, Anders HJ, Rudelius M, Schmidt-Supprian M. (2020). c-Rel gain in B cells drives germinal center reactions and autoantibody production. J Clin Invest. https://doi.org/10.1172/JCI124382 (link is external)
Overview
Pancreatic cancer is one of the biggest challenges for oncologists and scientists: It belongs to those forms of cancer that are very aggressive and difficult to treat. Every year around 450,000 people around the world receive the diagnosis of pancreatic cancer. Survival in this cancer is still the lowest among all cancers: The overall 5-year survival rate is less than 8%. This has remained almost unchanged over the last 30 years, despite tremendous efforts in preclinical and clinical science. Pancreatic cancer is predicted to become the second leading cause of cancer death in the next decade.
The molecular and cellular mechanisms linking the immune system with pro-tumorigenic signaling pathways in PDAC cells and their microenvironment are poorly defined. We aim to systematically dissect inflammatory signals derived from defined tumor subtypes and their microenvironment and investigate how these signals influence PDAC progression, therapeutic efficacy and treatment resistance. With our experiments, we aim to uncover targetable inflammatory drivers and develop novel treatment strategies for translation into the clinic in the future.
More information: https://sfb1321.med.tum.de/en(link is external)
